Using generative models as a bridge to understand the biological and artificial intelligence.
My goal is to use the first principle of intelligence to bridge the gap between artificial and biological intelligence. This theory of closed-loop transcription via rate reduction and information theory can help build a generative model that performs well on image synthesis and novelty detection. Furthermore, the generative model can enhance the visual cortex computation because it shares the same property that the human or primate visual cortex's abstract attribute resembles the generative model's latent attribute. Since memory is associated with visual stimuli, the generative model can investigate the relationship between the hippocampus and the visual cortex. Additionally, the insights from memory can be applied to guide continual learning. Finally, I want to apply this new technology to solve problems in science (e.g., biology, chemistry, and physics).
Generative model, Visual cortex computation, Memory modelling, Continual learning, AI for Science.
Research Interests
- Generative model(e.g., VAE, PixelCNN, GAN, Glow, and Diffusion model) via first priciple of intellgince (e.g., Information theory, Closed-loop Transcription, and Reference prior )
- Visual cortex computation via Generative model(e.g.,Visual cortex modeling, Neural encoding and decoding)
- Memory modelling with Visual cortex computation (Hippocampus modelling)
- Continual learning via memory modelling (e.g., Excitation-inhibition balance, Learning method of NN, Self-supervised learning)
- Application of generative model with Continual learning (e.g., Novelty detection, Image synthesis, AI for Science (e.g.,Structural variants detection(computational biology), Catalytic reaction(Chemistry))
Projects
Generative model for out-of-distribution detection and image synthesis
- VAE for out-of-distribution detection
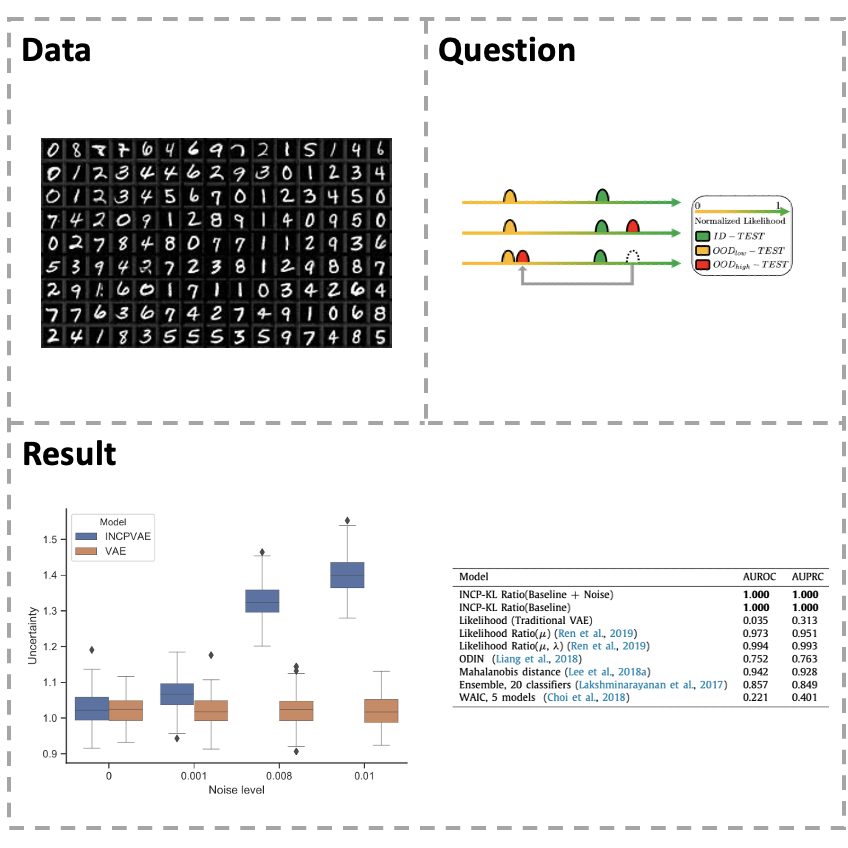
[Paper] Variational autoencoders (VAEs) are influential generative models with rich representation capabilities from the deep neural network architecture and Bayesian method. However, VAE models have a weakness that assign a higher likelihood to out-of-distribution (OOD) inputs than in-distribution (ID) inputs. To address this problem, a reliable uncertainty estimation is considered to be critical for in- depth understanding of OOD inputs. In this study, we propose an improved noise contrastive prior (INCP) to be able to integrate into the encoder of VAEs, called INCPVAE. INCP is scalable, trainable and compatible with VAEs, and it also adopts the merits from the INCP for uncertainty estimation. Experiments on various datasets demonstrate that compared to the standard VAEs, our model is superior in uncertainty estimation for the OOD data and is robust in anomaly detection tasks. The INCPVAE model obtains reliable uncertainty estimation for OOD inputs and solves the OOD problem in VAE models.
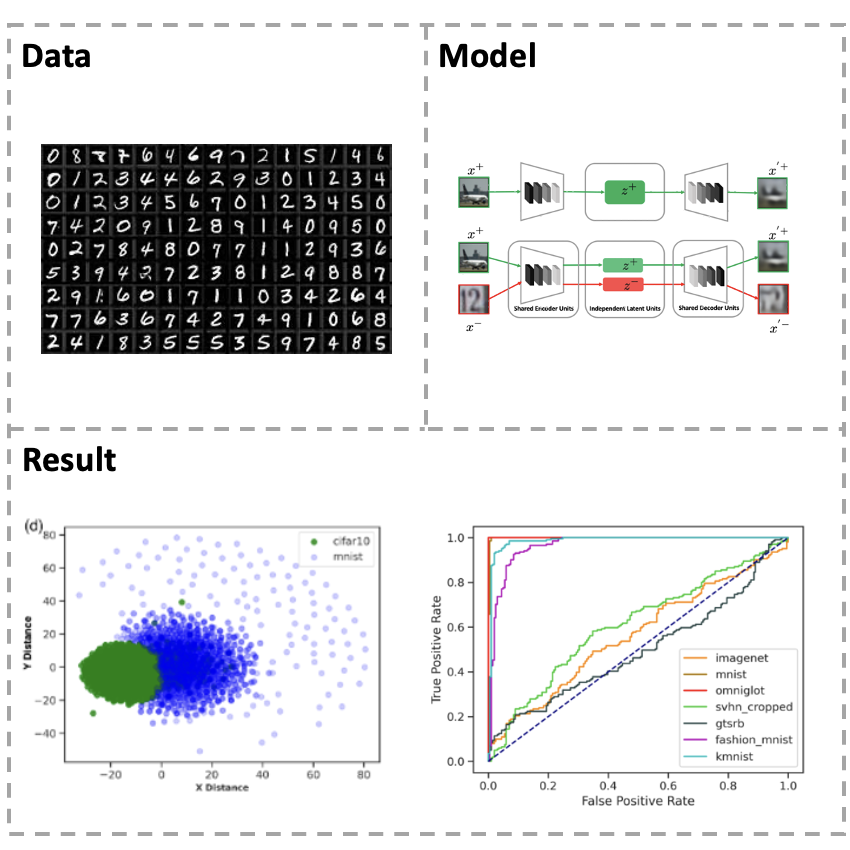
[Paper] We study the out-of-distribution detection problem using likelihood-based models. We propose a novel \textit{Bigeminal Priors Variational Autoencoder (BPVAEs)} method to remediate the possible higher likelihood of VAEs on the out-of-distribution (OOD) examples than on the in-distribution (ID) examples. With shared encoder and decoder networks, our BPVAE models the ID and OOD data simultaneously using ID priors and OOD priors respectively. We prove that, given the appropriate priors, the OOD data that would not have been detected using the single-prior VAEs model is guaranteed to have lower likelihoods than the ID data using our BPVAE model. We also show empirically that our BPVAE model can generalize to the unseen OOD examples. We compare our model with other likelihood-based OOD detection methods. Our numerical results demonstrate the effectiveness of BPVAEs on several benchmark OOD detection tasks.
- GAN for text-to-image synthesis
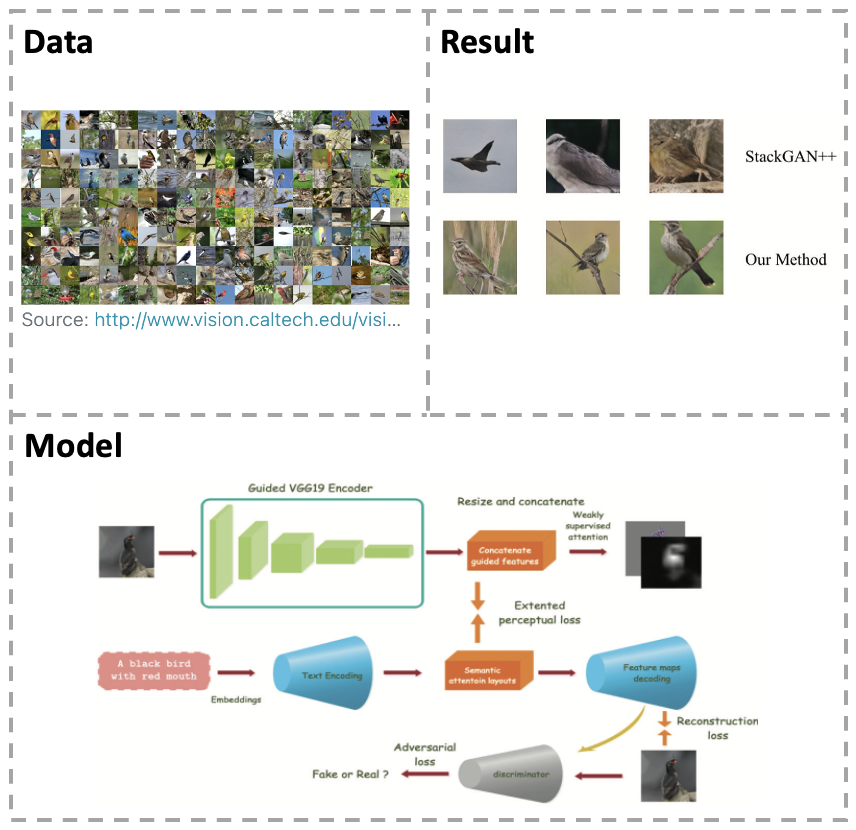
[Paper] Text-to-Image is a significant problem in computer vision. Question: Recently, there are some problems in the quality and semantic consistency of the generated image. Method:In this paper we propose an approach for Text-to-Image synthesis by focusing on the perception. We use text embeddings to generate semantic feature maps before target images synthesis instead of generating target images directly. The ground truth semantic layouts are calculated by interpretable classification network, and we will learn to generate semantic layouts before inferring target images from them. Result:We have trained our approach on the CUB2011 dataset and verified the quality of its generation and the interpretability of the network in simple background and small scale feature generation.
- GAN for medical image synthesis
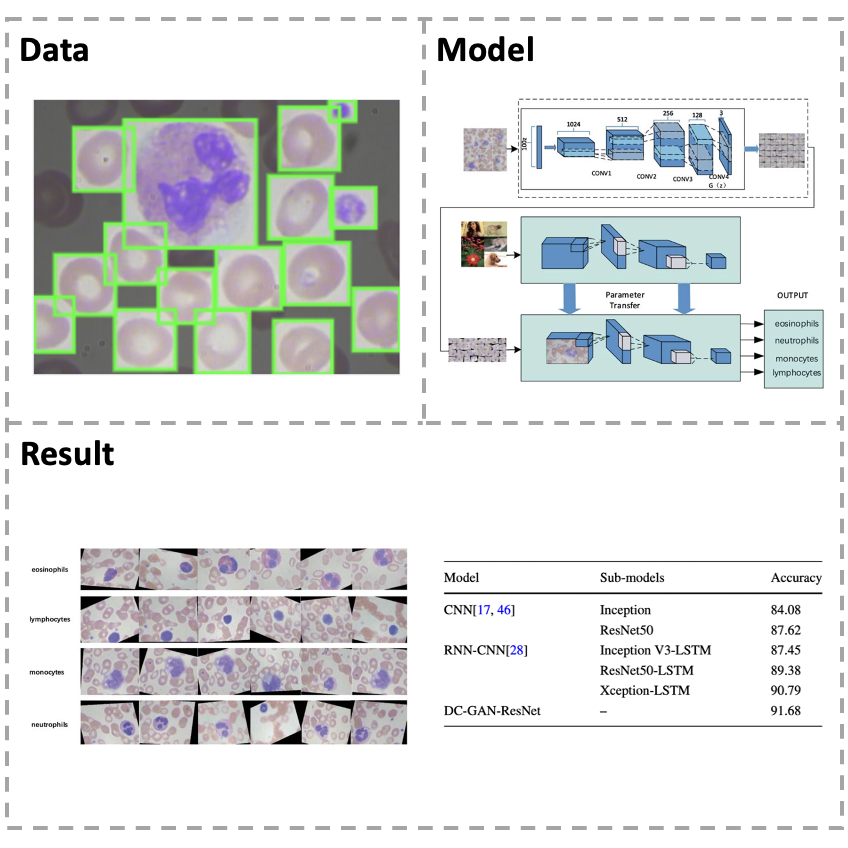
[Paper] In medicine, white blood cells (WBCs) play an important role in the human immune system. The different types of WBC abnormalities are related to different diseases so that the total number and classification of WBCs are critical for clinical diagnosis and therapy. However, the traditional method of white blood cell classification is to segment the cells, extract features, and then classify them. Such method depends on the good segmentation, and the accuracy is not high. Moreover, the insufficient data or unbalanced samples can cause the low classification accuracy of model by using deep learning in medical diagnosis. To solve these problems, this paper proposes a new blood cell image classification framework which is based on a deep convolutional generative adversarial network (DC-GAN) and a residual neural network (ResNet). In particular, we introduce a new loss function which is improved the discriminative power of the deeply learned features. The experiments show that our model has a good performance on the classification of WBC images, and the accuracy reaches 91.7%.
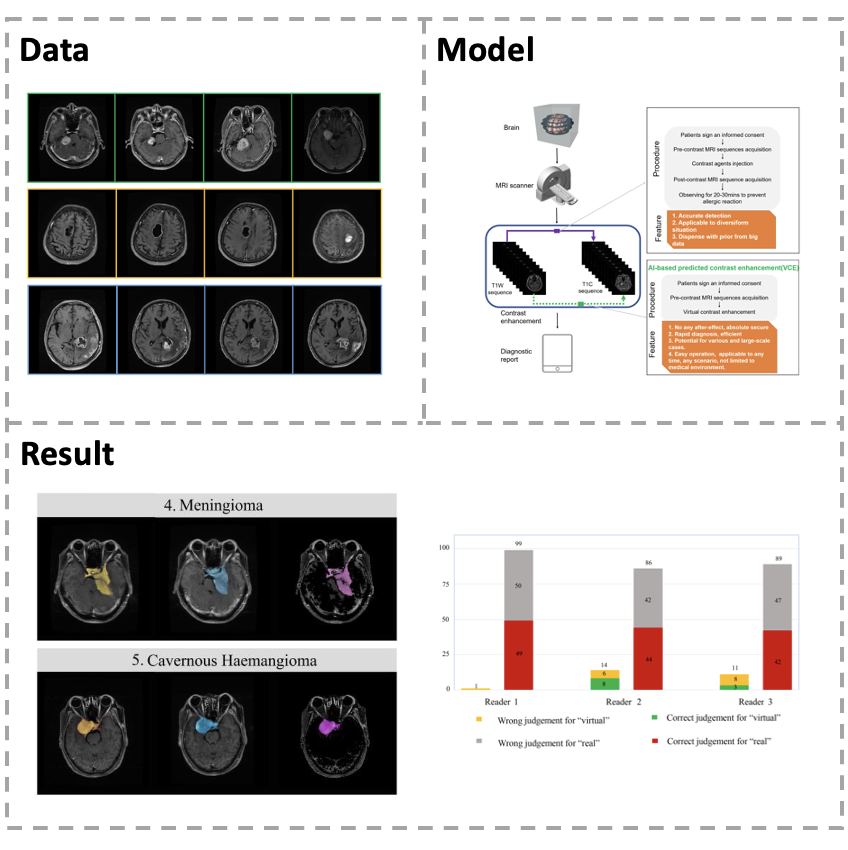
[Paper] Gadolinium-based-contrast-agents (GBCAs) injection has some potential risks for occurrence of adverse events and gadolinium deposition in brain tissue. We collected a dataset containing 113060 pairs of precontrast T1-weighted images and contrast-enhanced T1-weighted images (T1C) including abundant lesion categories to develop a novel virtual contrast enhancement model. Our model can utilize long-dependencies and peripheral information to acquire precise mapping transformation via logical inferences. Large-scale studies with qualitative evaluation and quantitative measurement indicate that our proposed model can not only generate virtual MR T1C with high fidelity, but also produce accurate enhancement results in both normal structures and intracranial lesions.
Visual cortex modelling and neural encoding and decoding
- Visual cortex modelling
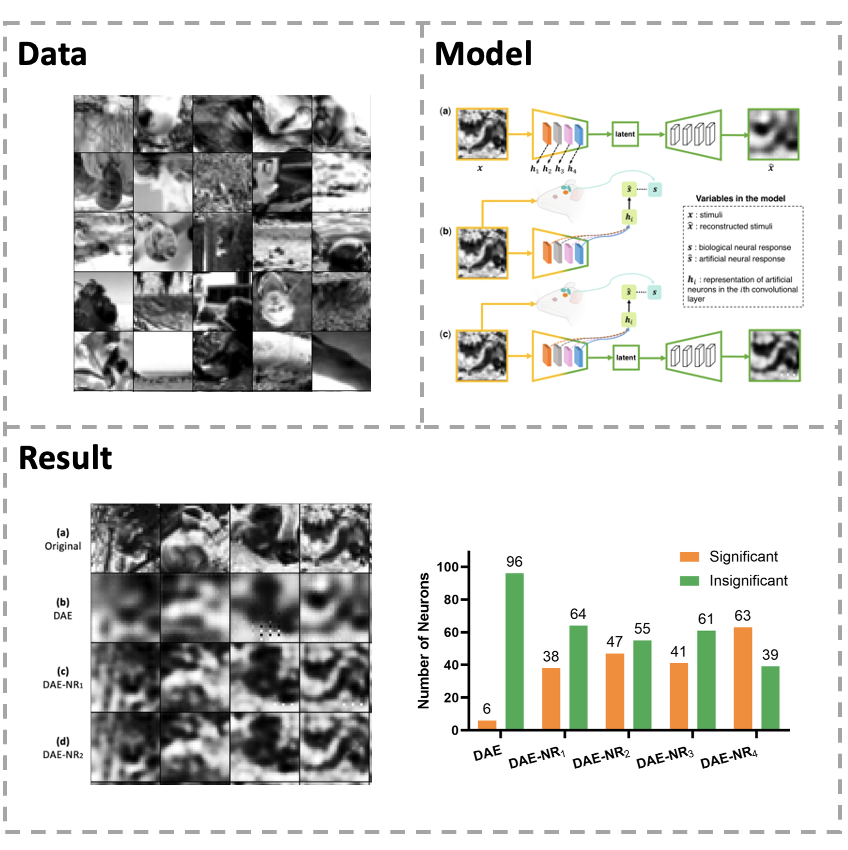
[Paper] Artificial neural network (ANN) is a versatile tool to study the neural representation in the ventral visual stream, and the knowledge in neuroscience in return inspires ANN models to improve performance in the task. However, it is still unclear how to merge these two directions into a unified framework. In this study, we propose an integrated framework called Deep Autoencoder with Neural Response (DAE-NR), which incorporates information from ANN and the visual cortex to achieve better image reconstruction performance and higher neural representation similarity between biological and artificial neurons. The same visual stimuli (i.e., natural images) are input to both the mice brain and DAE-NR. The encoder of DAE-NR jointly learns the dependencies from neural spike encoding and image reconstruction. For the neural spike encoding task, the features derived from a specific hidden layer of the encoder are transformed by a mapping function to predict the ground-truth neural response under the constraint of image reconstruction. Simultaneously, for the image reconstruction task, the latent representation obtained by the encoder is assigned to a decoder to restore the original image under the guidance of neural information. In DAE-NR, the learning process of encoder, mapping function and decoder are all implicitly constrained by these two tasks. Our experiments demonstrate that if and only if with the joint learning, DAE-NRs can improve the performance of visual image reconstruction and increase the representation similarity between biological neurons and artificial neurons. The DAE-NR offers a new perspective on the integration of computer vision and neuroscience.
- Neural encoding and decoding
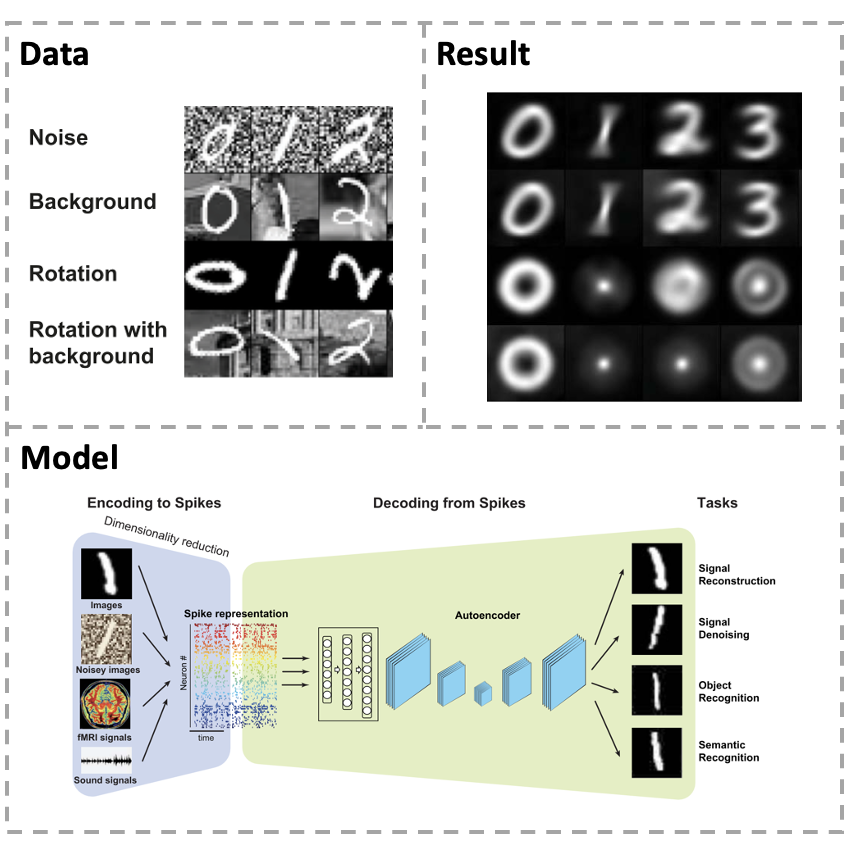
[Paper] Neural coding, including encoding and decoding, is one of the key problems in neuroscience for understanding how the brain uses neural signals to relate sensory perception and motor behaviors with neural systems. However, most of the existed studies only aim at dealing with the continuous signal of neural systems, while lacking a unique feature of biological neurons, termed spike, which is the fundamental information unit for neural computation as well as a building block for brain–machine interface. Aiming at these limitations, we propose a transcoding framework to encode multi-modal sensory information into neural spikes and then reconstruct stimuli from spikes. Sensory information can be compressed into 10% in terms of neural spikes, yet re-extract 100% of information by reconstruction. Our framework can not only fea- sibly and accurately reconstruct dynamical visual and auditory scenes, but also rebuild the stimulus patterns from functional magnetic resonance imaging (fMRI) brain activities. More impor- tantly, it has a superb ability of noise immunity for various types of artificial noises and background signals. The proposed framework provides efficient ways to perform multimodal feature representation and reconstruction in a high-throughput fashion, with potential usage for efficient neuromorphic computing in a noisy environment.
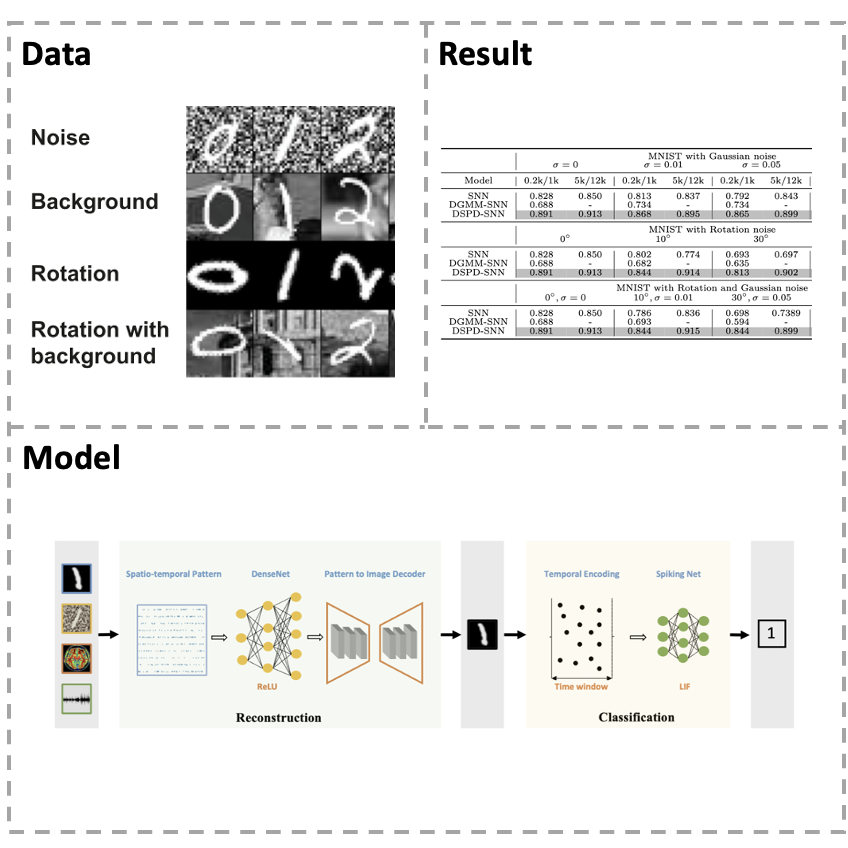
[Paper] Sensory information recognition is mainly processed through ventral and dorsal visual pathways in the primate brain visual system, which has layered and feature representations that show a strong resemblance to convolutional neural networks (CNNs), including reconstruction and classification. However, most of the existing studies treat them as a separate part, either considering only pattern reconstruction or classification tasks, without considering a particular feature of biological neurons, which are the critical visual sensory information units to neural computation. To solve these problems, we propose a uniform framework for sensory information recognition with augmented spikes. By integrating pattern reconstruction and classification into one framework, the framework can reasonably and precisely reconstruct multi-modal sensory information but also classify them by giving final label answers. The conducted experiments on video scenes, static image datasets, dynamic auditory scenes, and functional magnetic resonance imaging (fMRI) brain activities demonstrate that our framework delivers state-of-the-art pattern reconstruction quality and classification accuracy. Our framework enhances biological realism in multi-modal pattern recognition models. We hope to shed light on how the primate brain visual system performs this reconstruction and classification task by combining ventral and dorsal pathways.
Learning method for training Neural Network
- Hybrid learning with inhibition-excitation mechanism for training SNN
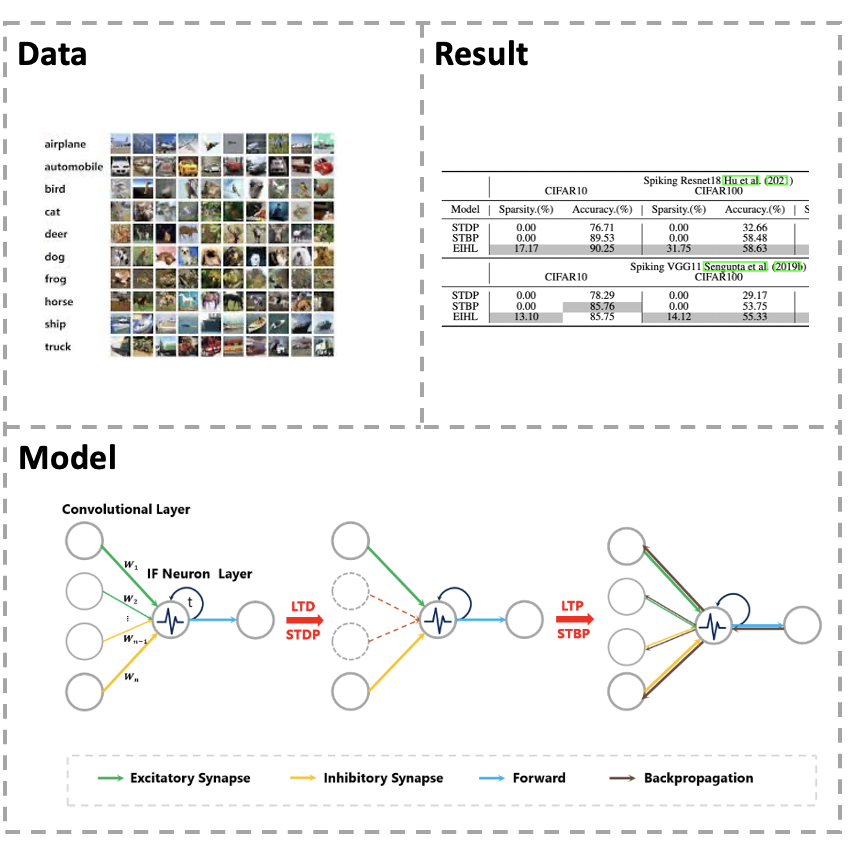
[Paper] The training method of Spiking Neural Networks (SNNs) is an essential problem,and how to integrate local and global learning is a worthy research interest. However, the current integration methods do not consider the network conditions suitable for local and global learning and thus fail to balance their advantages. In this paper, we propose an Excitation-Inhibition Mechanism-assisted Hybrid Learning (EIHL) algorithm that adjusts the network connectivity by using the excitation-inhibition mechanism and then switches between local and global learning according to the network connectivity. The experimental results on CIFAR10/100 and DVS-CIFAR10 demonstrate that the EIHL not only obtains better accuracy performance than other methods but also has excellent sparsity advantage. Especially, the Spiking VGG11 is trained by EIHL, STBP, and STDP on DVS CIFAR10, respectively. The accuracy of the Spiking VGG11 model with EIHL is 62.45%, which is 4.35% higher than STBP and 11.40% higher than STDP. Furthermore, he sparsity achieves 18.74%, which is quite higher than the above two non-sparse methods. Moreover, the excitation-inhibition mechanism used in our method also offers a new perspective on the field of SNN learning.
AI for Science
- Multi-MAE for structural variants detection
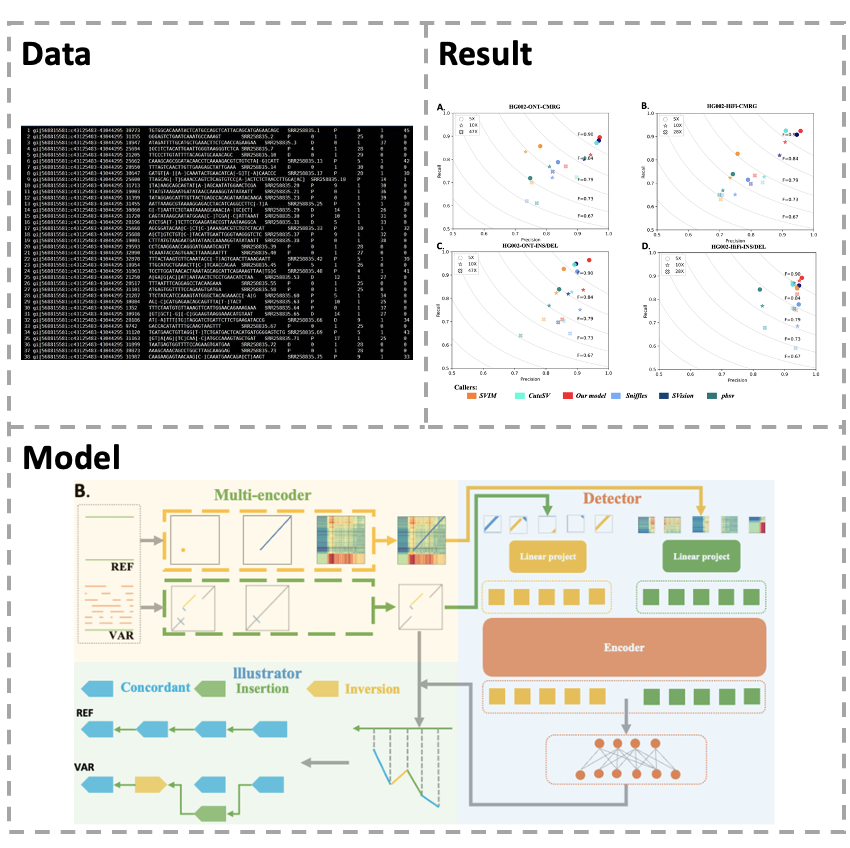
The detection of human genomic structural variants (SVs) is essential for studying genetic evolution and disease progression. Recent advances in long-read sequencing technologies have greatly facilitate SV detection, and the rich SV features derived from long reads even enabled complex SV discovery.
To get better performance for SV detection, we developed GeneMAE to detect SVs from long reads.
Inspired by multi-modal learning, here we convert the detection of SVs into a targeted multi-object recognition task, using two modalities of images to represent the SV feature sequences and a deep neural network to detect different types of SVs from the modalities.
Specifically, the multi-modal framework leverages two distinct modalities of SV: SVision and Cue. We demonstrate that GeneMAE can exploit the information sharing between modalities to capture more features of SVs and achieve higher accuracy for both simple and complex SVs.
This study provides a multi-modal perspective for SV detection and demonstrates the potential of applying deep learning in genomics.
Publications
- Xuming Ran, Jie Zhang, Ziyuan Ye, Haiyan Wu, Qi Xu, Huihui Zhou, and Quanying Liu. A computational framework to unify representation similarity and function in biological and artificial neural networks. Under Review (Rivsion 1) at: IEEE Transactions on Pattern Analysis and Machine Intelligence, 2022.
- Xuming Ran, Mingkun Xu, Qi Xu, Huihui Zhou, and Quanying Liu. Bigeminal Priors Variational auto-encoder. arXiv preprint arXiv:2010.01819, 2020.
- Xuming Ran, Mingkun Xu, Lingrui Mei, Qi Xu, and Quanying Liu. Detecting out-of-distribution samples via variational auto-encoder with reliable uncertainty estimation Neural Networks, 2021.
- Jie Yuan, Xuming Ran, Keyin Liu, Chen Yao, Yi Yao, Haiyan Wu, and Quanying Liu. Machine Learning Applications on Neuroimaging for Diagnosis and Prognosis of Epilepsy: A Review, Journal of neuroscience methods, 2021 .
- Lingrui Mei, Xuming Ran, and Jin Hu. Weakly Supervised Attention Inference Generative Adversarial Network for Text-to-Image, 2019 IEEE Symposium Series on Computational Intelligence (SSCI), 2019.
- Qi Xu, Jiangrong Shen, Xuming Ran, Huajin Tang, Gang Pan, and Jian K. Liu. Robust transcoding sensory information with neural spikes, IEEE Transactions on Neural Networks and Learning Systems, 2021.
- Li Ma, Renjun Shuai, Xuming Ran, Wenjia Liu, and Chao Ye. Combining DC-GAN with ResNet for blood cell image classification, Medical & biological engineering & computing 58, no. 6 :1251-1264, 2020.
- Qi Xu, Yuyuan Gao, Jiangrong Shen, Yaxin Li, Xuming Ran, Huajin Tang, Gang Pan, Enhancing Adaptive History Reserving by Spiking Convolutional Block Attention Module in Recurrent Neural Networks, NeurIPS, 2023.
- Shan-Shan Li, Yu-Shi Jiang, Xue-Ling Luo, Xuming Ran, Yuqiang Li, Dong Wu, Cheng-Xue Pan, Peng-Ju Xia, Photocatalytic Vinyl Radical-Mediated Multicomponent 1,4-/1,8-carboimination Across Alkynes and Olefins/(Hetero)Arenes, Science China Chemistry , 2023.
- Songming Zhang, Xiaofeng Chen, Xuming Ran, Zhongshan Li, Wenming Cao, Even decision tree needs causality, Under Review at: IEEE Transactions on Neural Networks and Learning Systems, 2022.
- Hong Peng, Mingkun Xu Bo Wang, Zheyu Yang, Xuming Ran, Bo Li, Jiaohua Huo, Jing Pei, Yuanyuan Cui , Huafeng Xiao, Xin Lou, Cuiping Mao, Guangming Zhu, Liang zhang , Zheng You, Lin Ma, A New Virtual MR Contrast-enhancement Method based on Deep Learning: Faster, Safer, and Easier, Under Review at: Nature Machine Intelligence, 2022.
- Qi Xu, Sibo Liu, Xuming Ran, Yaxin Li, Jiangrong Shen, Huajin Tang, Jian K. Liu, and Gang Pan, Robust Sensory Information Reconstruction and Classification with Augmented Spikes, Under Review at: IEEE Transactions on Neural Networks and Learning Systems, 2023.
- Tingting Jiang, Qi Xu, Xuming Ran, Jiangrong Shen, Pan Lv, Qiang Zhang, Gang Pan, Adaptive deep spiking neural network with global-local learning via balanced excitatory and inhibitory mechanism, ICLR, 2023.
- Mengyu Yang, Ye Tian, Rui Su, Xuming Ran, ViMoV2: Efficient Recognition for Long-untrimmed Videos with Multi-modalities, Under Review at: AAAI, 2023
Talks
Professional Service
Conference Reviewing
Journal Reviewing
Summer School